
Chemical Proteomics
Evotec scientists have pioneered chemical proteomics applications supporting and driving discovery of new drugs across the full value chain. These include methods for deconvoluting targets of hits from phenotypic screens, investigating the selectivity and target profile of drug candidates, determining the mechanism-of-action of small molecules. In addition, methods have been developed to chemically label and identify novel sites in proteins for drug interactions.
Target Deconvolution
Evotec has elaborated chemical proteomics applications to support target deconvolution of bioactive compounds emerging from phenotypic screens. Cellular target identification is the crucial step to enable further drug optimisation and development.
Evotec Cellular Target Profiling™ is a powerful technology to identify specific cellular compound targets including affinity constants in lysates from any cell type or tissue. Evotec’s chemical proteomics offering further comprises photoaffinity labelling (PAL-MS) for covalent target capture in live cells.
Our offering comprises:
- Evotec’s chemical proteomics approaches use high-end quantitative mass spectrometry to reveal and verify specific cellular targets of a drug
- Evotec Cellular Target Profiling™ (CTP) determines target-specific dissociation constants for the compound studied, ranking targets according to their likely physiological relevance
- Drug photoaffinity labelling (PAL-MS) allows target identification in live cells, and supports binding site identification in protein targets.
- Unbiased target ID by proteome-wide profiling of native, endogenously expressed, post-translationally modified proteins in the presence of cellular co-factors and endogenous complex partners
- Complementary chemical proteomics approaches can be performed in an integrated fashion
- Extensive, non-target class restricted track record in successful profiling of diverse small molecule compounds
- CTP and PAL-MS approaches require derivatization of the small molecule of interest which can be covered by a dedicated team of chemists at Evotec.
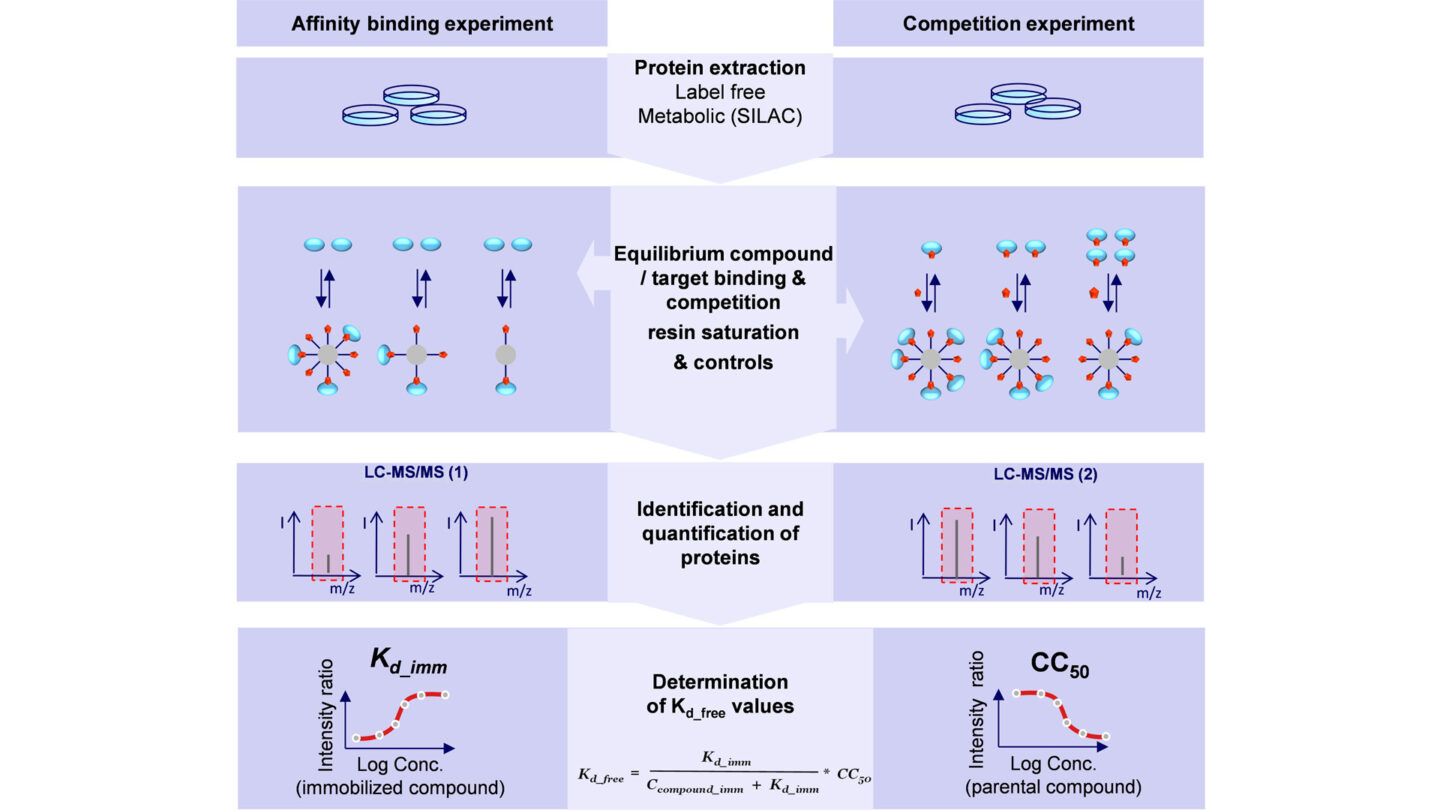
Cellular Target Profiling (CTP) Workflow
- Identification of small molecule targets in any type of cell of tissue of choice
- Determination of compound's proteome wide binding affinities (Kd values)
- Profiling of a compound against native, endogenously expressed, post-translationally modified full length proteins in the presence of cellular cofactors and native complex partners
- CTP requires a linker derivative of the active compound for target enrichment by affinity chromatography
- Synthesis of linker compounds with terminal NH2, COOH, of alkyne, azide moiety for functional immobilization
- Competition with active parental and inactive analogous compound
- CTP as mature "workhorse" approach for target ID with extensive, non-target class restricted track record in target deconvolution and profiling of various small molecules
Global Assessment of Drug Target Engagement & Selectivity
Evotec has established a chemical proteomics platform which employs high-end quantitative mass spectrometry for global assessment of drug target engagement and selectivity in the context of endogenous proteomes and sub-proteomes. Selectivity data about cellular on- and off-target liabilities is particularly useful for informed decisions at various stages of drug development, for example in the lead optimization phase or in the pre-clinical candidate selection process.
Our offering comprises:
- Evotec Cellular Target Profiling™ for unbiased, proteome-wide selectivity profiling to identify and quantify compound interactions with cellular on- and off-targets
- KinAffinity® as Evotec’s hit-to-lead compatible approach for rapid target profiling of kinase inhibitors in cell and tissue samples. Unlike traditional biochemical kinase panel screening, the inhibitor’s target affinities are determined simultaneously for a large number of endogenous kinases within their physiological cellular environment
- Photoaffinity labeling has been expanded to map reversible protein-ligand interactions (“ligandability map”) directly in live cells
- Activity-based protein profiling (ABPP) for a wide range of enzyme classes including serine hydrolases, metalloproteases, oxidoreductases, histone deacetylases, and glutathione S-transferases. ABPP visualizes only the active forms of particular enzymes using reactive chemical probes directed to the active site of a particular target protein family. Finally, ABPP is the method of choice to profile reactive nucleophilic amino acid residues (cysteine, lysine, tyrosine, aspartate/glutamate, histidine, and methionine) at the global scale (“hotspot map”) and map proteome-wide reactivity and selectivity of electrophiles across molecular scaffolds (“reactivity map”)
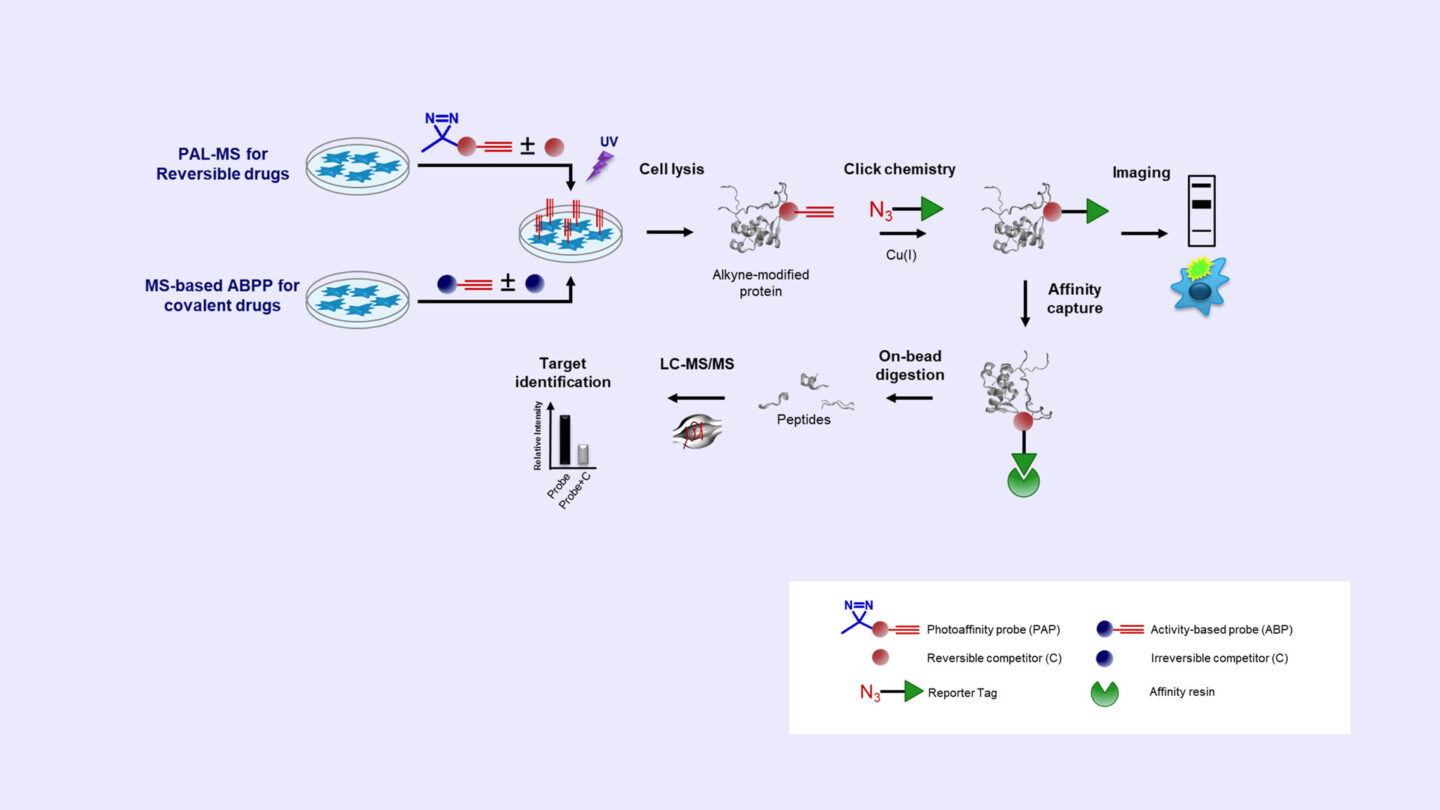
Photoaffinity Labeling Mass Spectrometry and Activity-based Protein Profiling (ABPP)
- Identification of direct binding targets (and off-targets) of reversible or covalent drugs in live cells
- Cellular localization of the drug-target complex using fluorescence imaging microsopy
Mechanism-of-Action Analysis
Elucidation the mechanism-of-action (MoA) of a drug candidate is an essential task to understand phenotypic effects, or potential side effects. There are multiple methods to address this topic, among which are several proteomic approaches that are offered by Evotec. Interaction analysis using affinity enrichment via antibodies and co-purification of complexed proteins is well established at Evotec. Moreover, proximity labelling based on biotin ligase fusions (e.g. TurboID, UltraID) can be used to identify stable and transient interactions. Finally, methods are in place to elucidate affected pathways by monitoring posttranslational modifications such as phosphorylation, ubiquitinations, acetylations, or arginine methylations.
Our offering comprises:
- Antibody coupling to beads, and workflows to immunoprecipitate endogenous target proteins or tagged proteins, including software tools to identify true interactors
- TurboID- and UltraID-based proximity labeling in a semi-automated mode using Agilent BRAVO system.
- PTM enrichment methods are established in semi-automated mode for proteome-wide phosphorylation sites, lysine-acetylations, lysine-ubiquitinations, and arginine methylations. A bioinformatics tool box is set up for differential expression analysis, functional analysis, pathway mapping, kinase activity and motif analysis.
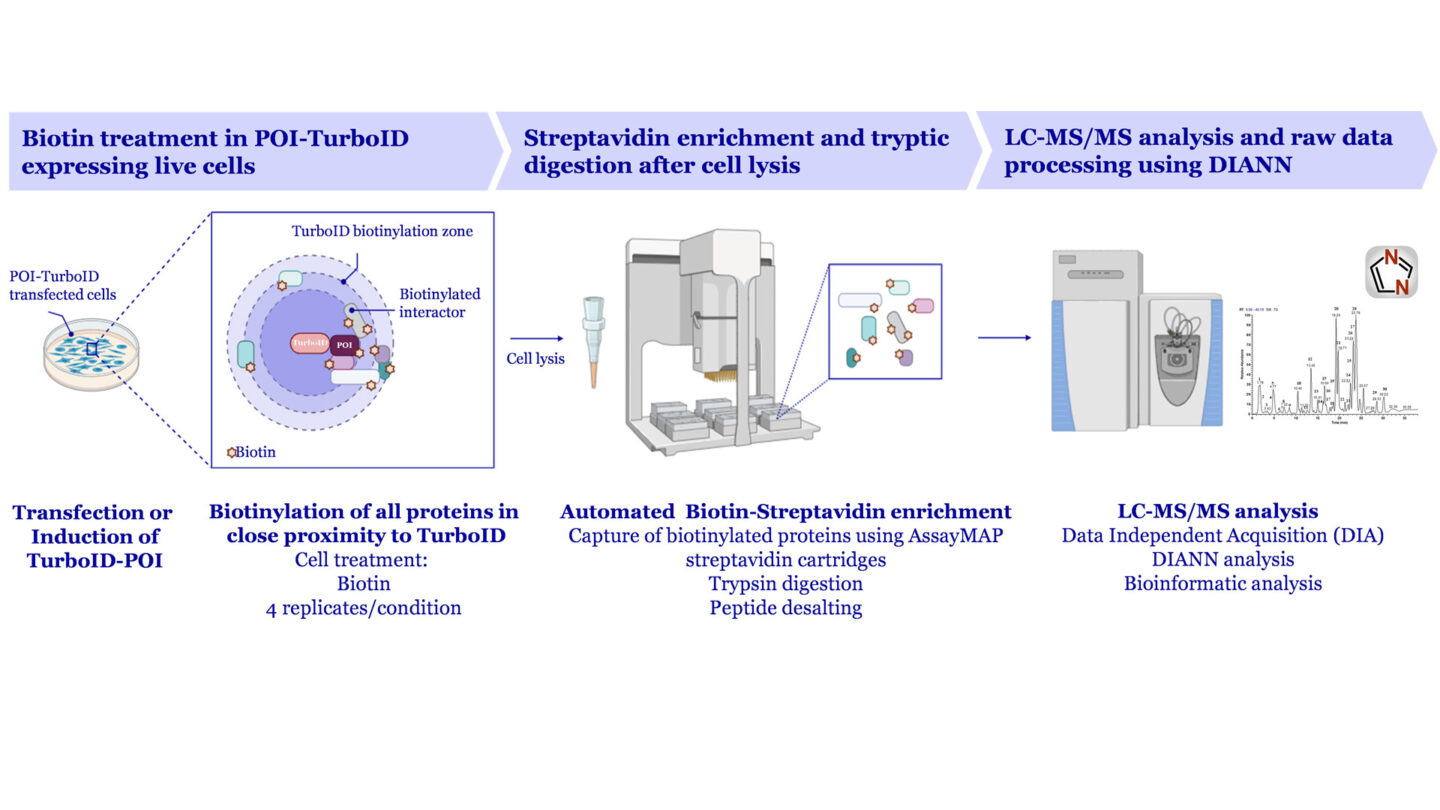
Proximity Labeling using TurboID
A semi-automated workflow allows a throughput of 20-40 conditions/month upon request with efforts to increase
- Proteome Profiling to accurately measure protein abundance levels on a global scale using Isotope labeling (SILAC), diMe, as well as label-free detection methods
- Deep Proteome Profiling with MS sample pre-fractionation or Single-shot Profiling for higher throughput
- Quantitative and global measurement of post-translational modification sites can be combined with proteome profiling
- Sales include cultured cells, fresh and FFPE tissues, subcellular fractions, biofluids from animals and humans, microorganisms, plants
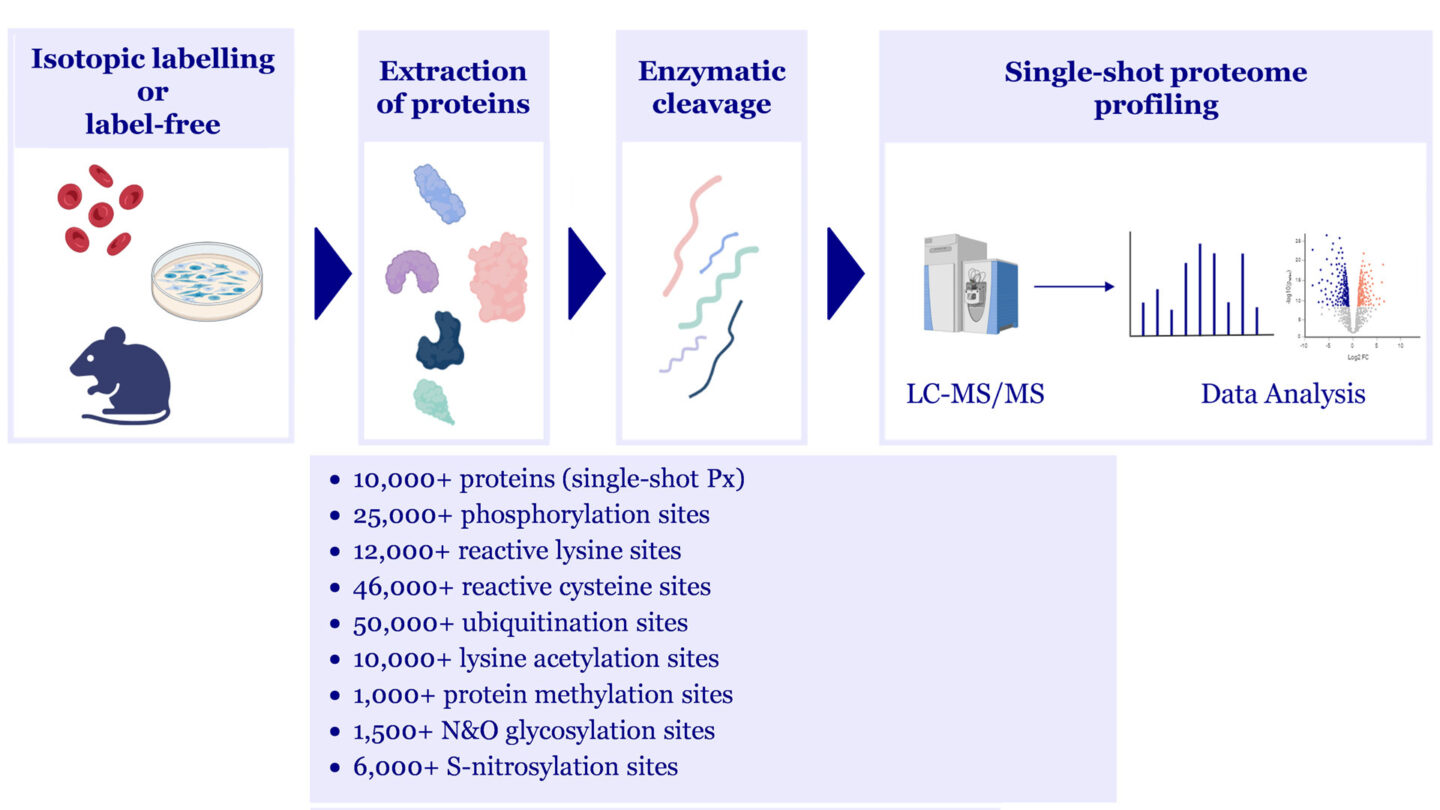
PTM analysis: workflow and capabilities
Typical project applications
- Drug mode-of-action analysis and pharmacodynamic markers
- Identification of disease-specific target and biomarker
- Biomarker discovery for patient stratification and efficacy-studies
- Disease comprehension
High Throughput Covalent Fragment-based Drug Discovery Platform
Evotec’s chemo-proteomic screening platform aims to discover unknown target sites within disordered regions, cryptic or shallow binding pockets, or allosteric pockets on the surface of proteins while simultaneously identifying small molecule fragments that interact in a highly selective manner with those pockets directly in complex biological systems (cell or tissue lysates, live cells, live organisms)
Proteomics can play a key role in the different stages of fragment-based drug discovery
- Target identification by looking in the proteome for ligandable / hotspot residues within a target of interest
- Peptide mapping to identify the residue covalently modified by the fragment ligand
- Target engagement & Selectivity profiling in cells or in animal models
Our High-Throughput Activity-Based Protein Profiling (HT-ABPP) platform leverages residue-based probes (RBPs) that possess broadly reactive covalent warheads developed to modify side chains on the targeted amino acid residue, including cysteine, lysine, tyrosine, and aspartate/glutamate residues. RBPs also bear a reporter group, such as a fluorophore (i.e. TAMRA) or an affinity tag (i.e. desthiobiotin) for identification and enrichment purposes. Alternatively, a bioorthogonal chemical handle such as an alkyne or azide can be installed on the probe, allowing the attachment of reporter or affinity tags after the protein target conjugation step for subsequent click chemistry-enabled applications. Our HT-ABPP platform combines a robotic liquid handling platform for enrichment and peptide clean-up, stable isotope labeling by amino acids in cell culture (SILAC), and data-independent acquisition (DIA)-mass spectrometry (MS)-based proteomic analysis.. The platform allows the consistent mapping of 60,000+ reactive cysteine sites on 13,000+ unique proteins, and 12,000+ distinct reactive lysine sites on 3,500+ unique proteins, across many different human cell lines and primary cells with a throughput of up to 100 samples per day. Mapping of reactive tyrosine and Asp/Glu sites in various cell lines is currently in progress.
Our offering comprises:
- Build a Hotspot Map: build a proprietary druggable hotspot map (set of ligandable sites identified for >90% of the human proteome) based on disease systems. Combine cutting-edge chemoproteomics with machine-learning algorithms, modeling, simulation, and structural information to discover binding pockets (or druggable hotspots) within targets of interest involved in critical disease pathways.
- Build Reactivity Map: Map proteome-wide selectivity across molecular scaffolds. Classify, for a given protein, the relative reactivities and the binding potential of specific regions enabling the prioritization of particular targets and the design of effective screening strategies.
- Determine the desired chemical property space and reactivity profiles to focus on the most promising areas of chemical space.
- Build/improve proprietary covalent fragment libraries: Iterative data-driven and evolving library design (annotate frequent binders, dark chemical matter, target-selective molecular scaffolds).
- Accelerate lead generation: Convert a historically undruggable target into a tractable project that can be accelerated through LO and onward to the clinic.
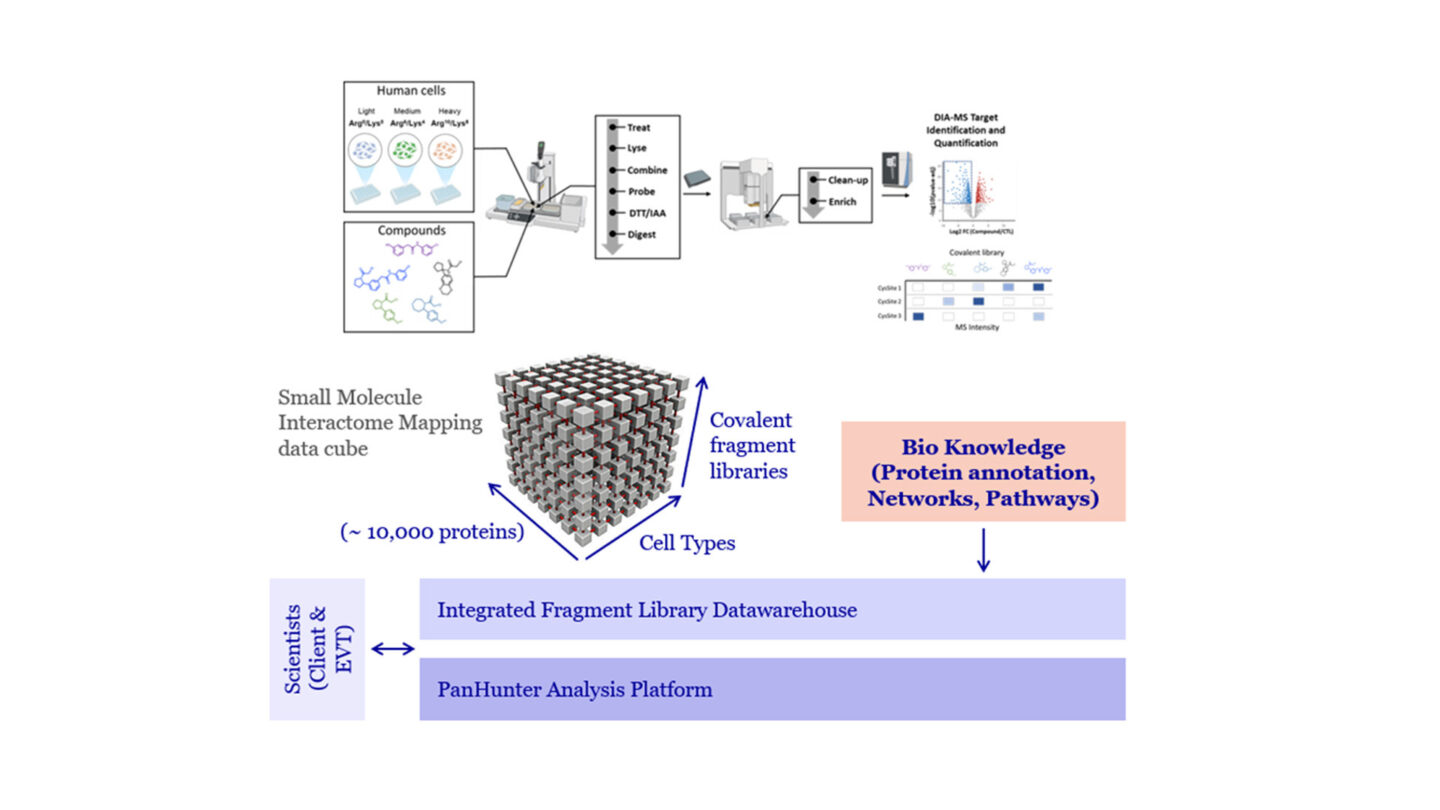
Overview of the HT-ABPP platform
Meet Our Experts
More on Our Science Pool
- Quantitative Proteomics for Target Deconvolution and Selectivity Profiling >
- https://sciencepool.evotec.com/quantitative-proteomics-for-target-deconvolution-and-selectivity-profiling/ >

