Understand the potential genotoxicity of your compound in our γH2AX double strand DNA damage response assay.
Our γH2AX double strand DNA damage response assay is in our portfolio of genotoxicity services. Cyprotex delivers consistent, high quality data and can adapt protocols based on specific customer requirements.
Introduction
Anti-γH2AX (phospho S139) as an indicator of DNA double-strand breaks (DSBs) and genotoxicity:
- Histone H2A variant H2AX, a component of the nucleosome core structure has a special role in DNA repair. Phosphorylation of H2AX at residue Ser-139 (Anti-γH2AX) by PI3K-like kinases, including ATM, ATR and DNA-PK, is an early cellular response to the generation of DNA double-strand breaks (DSBs)1.
- Detecting the formation of DSBs using the γH2AX (Ser-139) has emerged as a highly specific and sensitive molecular marker for monitoring DNA damage1.
- DSBs form when both strands of the DNA double helix are broken, irrespective of how they are formed they are found to be highly toxic and can ultimately be fatal2.
- Cyprotex’s DNA Damage assay uses High Content Screening (HCS) to identify both DNA Damage and Cytotoxicity.
Protocol
Double Strand DNA Damage Response Assay
Data
Data from Cyprotex's Double Strand DNA Damage Response Assay
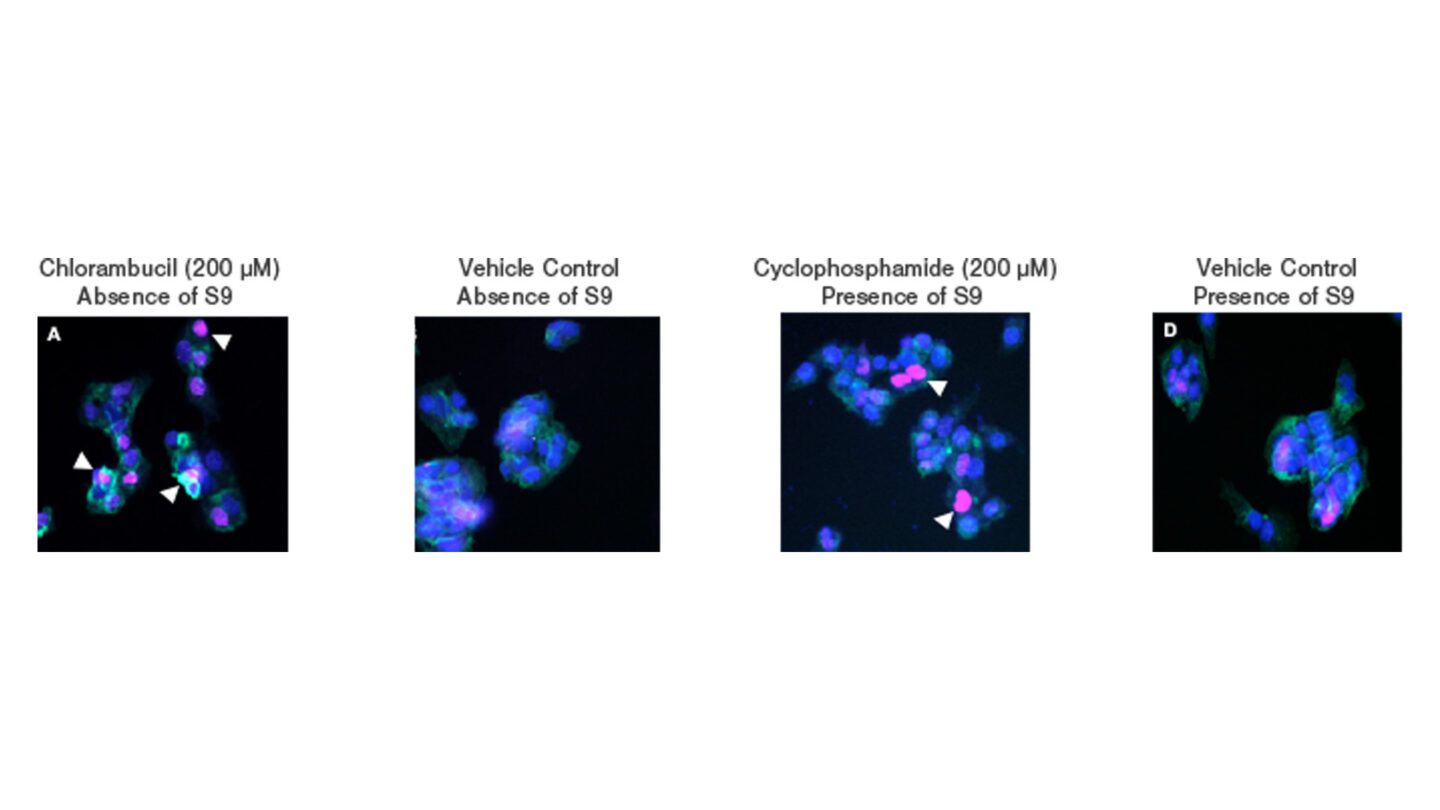
Figure 1
Representative HCS images for cells treated with (A) 200µM chlorambucil (positive control) (B) vehicle control in the absence of S9 fraction (C) 200µM cyclophosphamide (metabolising system positive control) and (D) vehicle control in the presence of S9 fraction over a 24 hr period. Cell nuclei are stained blue (Hoechst) with pink staining observed in the nucleus of cells positive for γH2A.X (indicated by white triangles). Cellular filamentous actin is stained green (phalloidin).
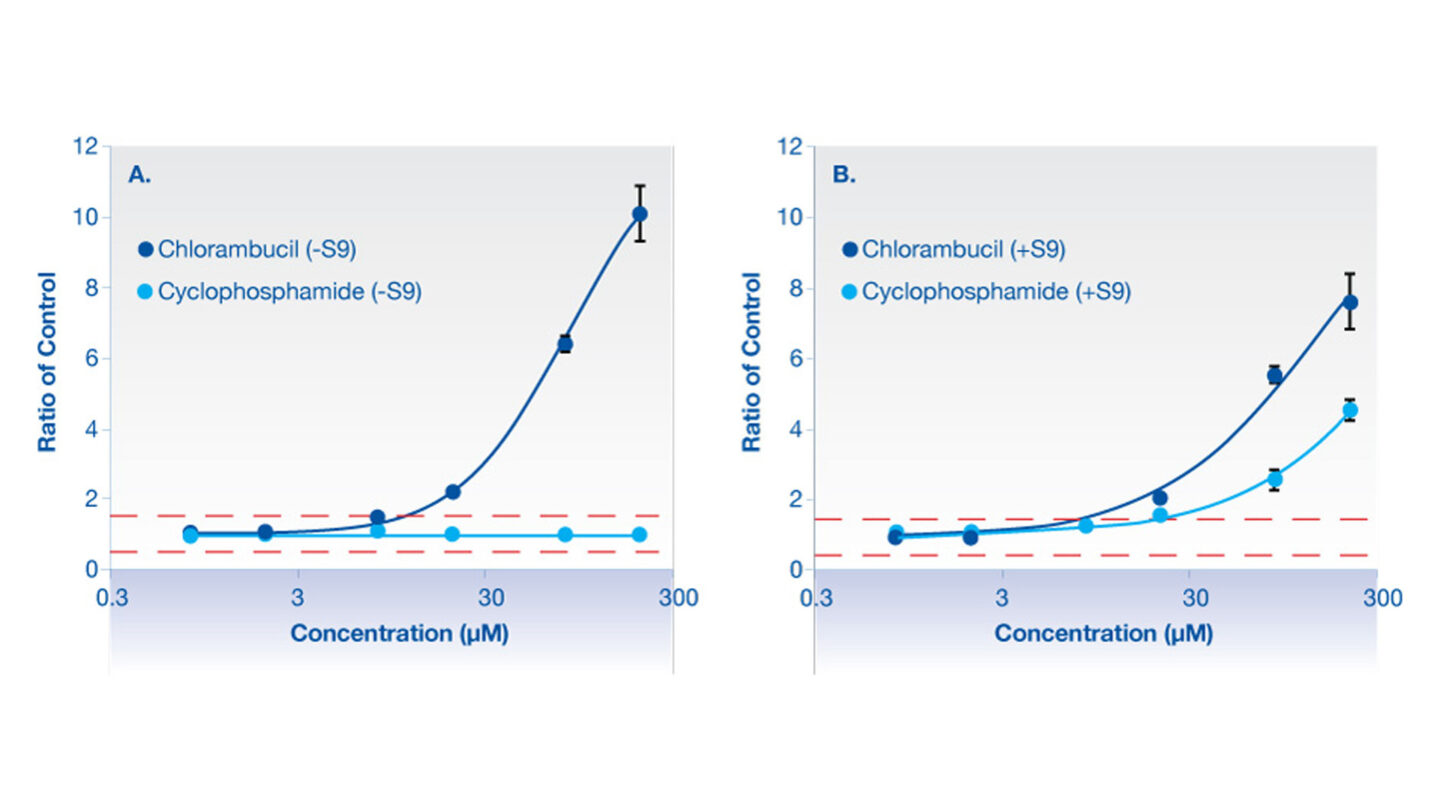
Figure 2
Graphical representation of γH2AX for chlorambucil (positive control) and cyclophosphamide (positive control for metabolizing system).
A: is in the absence of aroclor 1254 induced rat liver S9.
B: is in the presence of aroclor 1254 induced rat liver S9.
Red dashed line represents the vehicle control limits.
Chlorambucil causes a concentration dependent increase in γH2AX compared to vehicle control treated cells in both the absence and the presence of metabolizing system (aroclor 1254 induced rat liver S9). No response was observed for cyclophosphamide in the absence of metabolizing system, however in the presence of the metabolizing system cyclophosphamide shows a concentration dependent increase in γH2AX.
Data represents mean of triplicate incubations ± standard deviation.
References
1) Mah LJ et al., (2010) γH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia 24; 679 – 686
2) Chapman JR et al., (2012) Playing the End Game: DNA Double-Strand Break Repair Pathway Choice. Molecular Cell 47(4); 497-10
3) Tsamou M et al., (2012) Performance of in vitro γH2AX assay in HepG2 cells to predict in vivo genotoxicity. Mutagenesis 27(6); 645-652
4) Diaz D et al., (2007) Evaluation of an automated in vitro micronucleus assay in CHO-K1 cells. Mutation Research 630;1-13
5) Hastwell PW et al., (2009) Analysis of 75 marketed pharmaceuticals using the GADD45a-GFP ‘GreenScreen HC’ genotoxicity assay. Mutagenesis, 24(5); 455-463
Downloads
- Cyprotex Mechanisms of Drug Induced Toxicity Guide >
- Cyprotex γH2AX Double Strand DNA Damage Response Assay Factsheet >

